University of California, Berkeley | Plant & Microbial Biology
Pollen Proteomics
Why study pollen and sperm proteomics? That’s a great question. The process of double fertilization – the creation of the embryo and the endosperm in angiosperms – was discovered in 1898, but we still don’t know much about how the gametes recognize each other and fuse together. Why? This is mostly likely due to the fact that plant gametes are minuscule and embedded within other cells or tissues of the plant. With new techniques, we are now able to address some of the questions that have eluded us for years.
Recently, two studies of the Arabidopsis transcriptome (Honys & Twell, (2003) Plant Phys. 132 (2): 640-652 and Becker et al., (2003) Plant Phys. 133(2): 1-13) showed that about 20% of the 8200 genes analyzed (about 1/3 of the genome) were expressed both in pollen and in other tissues and about 10% of the genes had pollen-specific expression. We'd like to know when and where these mRNAs are translated into proteins. For example, some mRNAs might be translated only after pollen germination or upon gamete fusion and while others might be translated earlier in development. Thus, a proteomics approach will complement the pollen transcriptome analysis to give us a clearer picture of what’s going on.
Our approach focused on identifying salt insoluble proteins from mature Arabidopsis pollen. We believe that this protein fraction contains some of the membrane-bound and membrane–associated proteins that are likely to be involved in signaling during pollen tube growth and during fertilization.
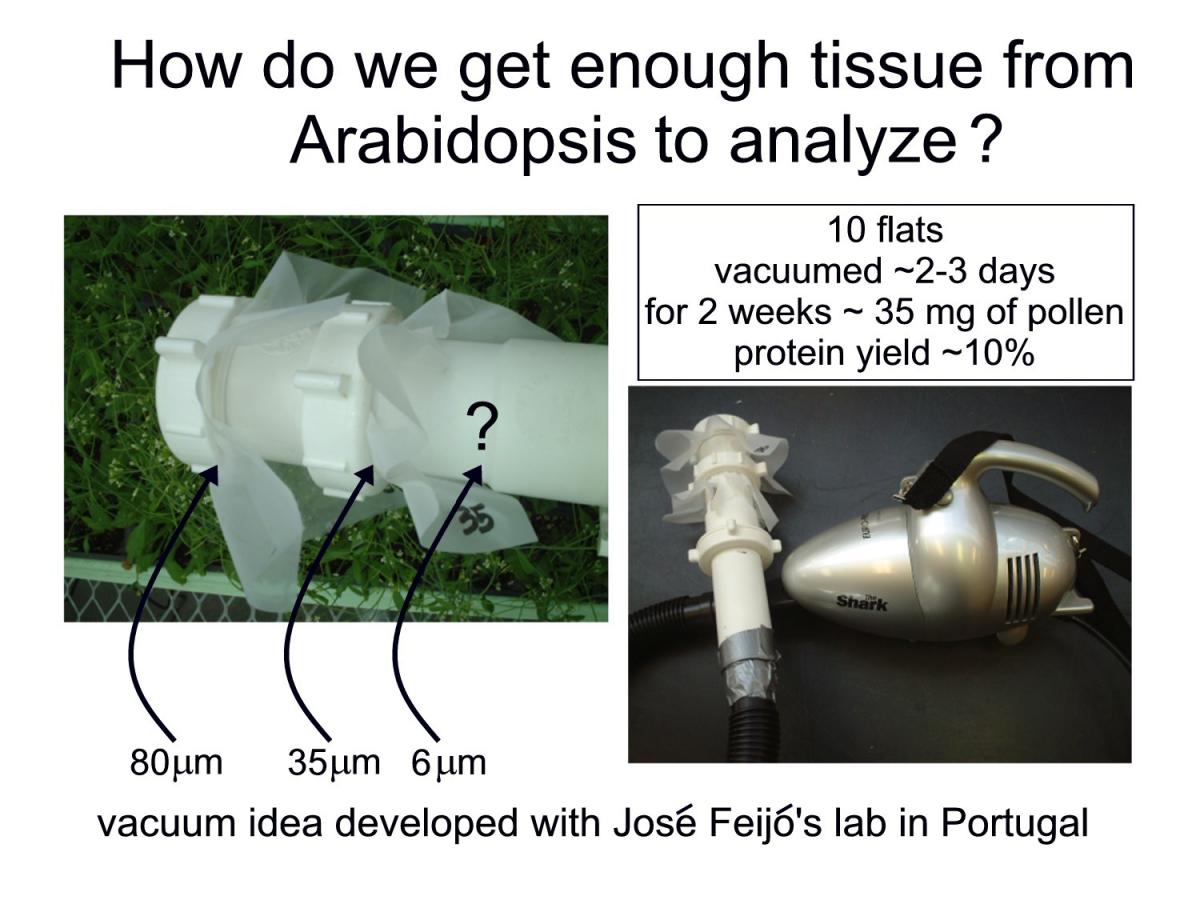
How are we doing this? Or, the more likely question is “How can you collect enough Arabidopsis pollen to do a proteomic analysis?” While Sheila McCormick was on sabbatical in José Feijo’s lab in Portugal, she took photographs of a household vacuum cleaner that researchers in the Feijo lab had adapted for pollen collection from Arabidopsis plants. Back in our lab, we modified the set-up using an inexpensive hand-held vacuum cleaner outfitted with common plumbing fittings, duct tape and nylon meshes of varying sizes. With the vacuum turned on, the “plumbing wand” is waved gently over the flowering plants. Flower parts and larger debris are collected on the outermost mesh and the pollen is collected on the inner smallest-sized mesh. This method works great! Dry pollen can be stored in the -80º C freezer until enough is accumulated for protein extraction. Details about how to vaccuum pollen can be found in Johnson-Brousseau and McCormick, 2004.
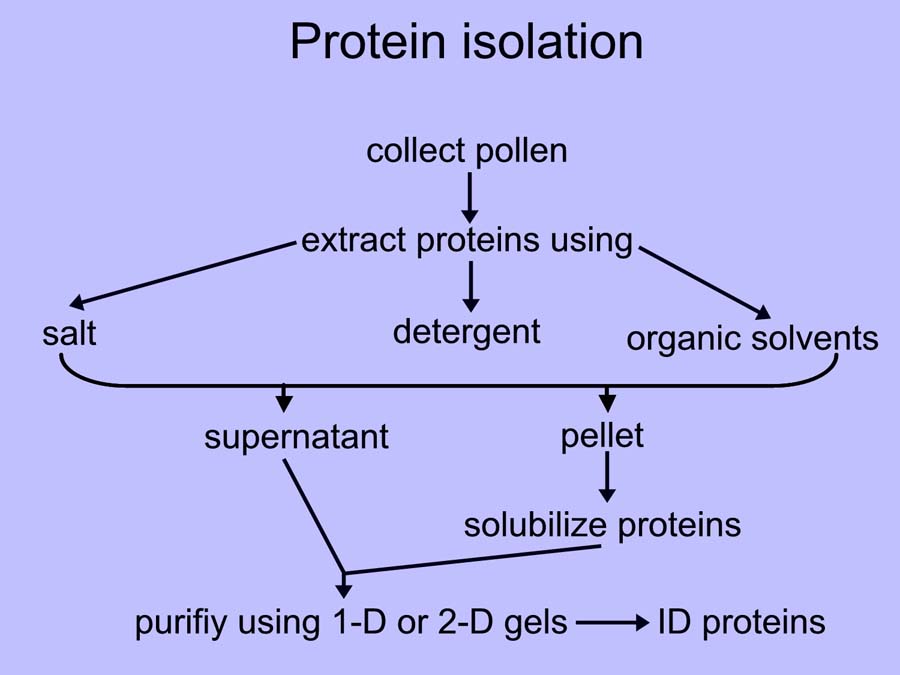
The protein isolation schemes we’ve tried are outlined in the figure below. Basically, the pollen is extracted with a high salt buffer and the resulting pellet is then reextracted with a buffer that contains detergents, chaotropes, reducing agents and ampholytes.
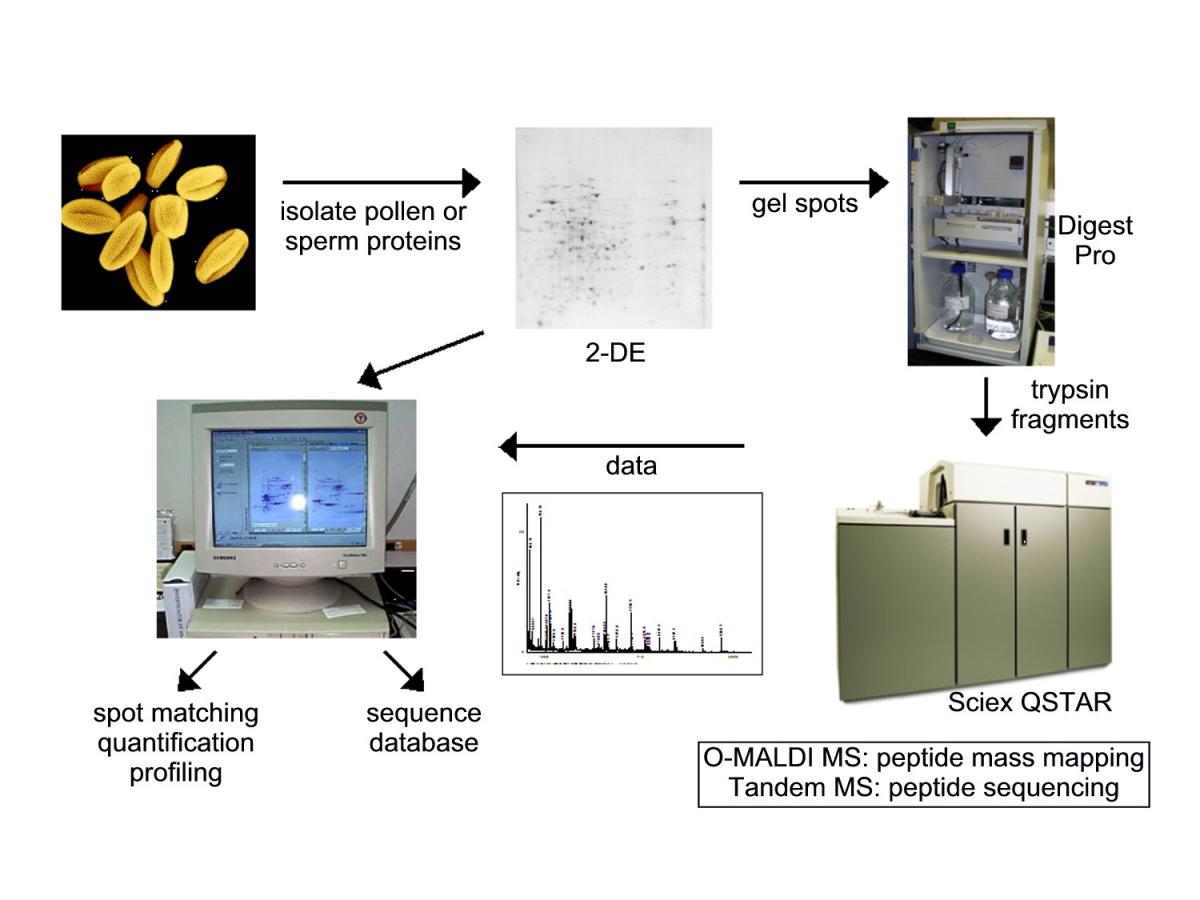
Then we walked our protein samples to the WRRC, on the other side of the building, where our collaborators, Bill Hurkman and Charlene Tanaka, ran the 2D gels. We then isolated the protein spots and Charlene digested them with trypsin using the automated Digest Pro system. The trypsinized peptide fragments were then subjected to either MALDI MS or Tandem MS by another one of our collaborators, Bill Vensel, and the data were collected and matched against the Arabidopsis database.
Collecting pollen and isolating proteins is one thing, but what about sperm? Another great question. Sperm cells are about 1% of the volume of a pollen grain. But by vacuuming a lot of Arabidopsis pollen, we think we we will be able to isolate sperm, using a transgenic line created in our lab that has GFP-tagged sperm. The fluorescent properties of the GFP should allow us to isolate pure samples of Arabidopsis sperm.
This work (Holmes-Davis, R., Tanaka, C.K., Vensel, W., Hurkman, W. and McCormick, S. 2005. Proteome mapping of mature pollen of Arabidopsis thaliana. Proteomics, Published Online: 24 Oct 2005, DOI: 10.1002/pmic.200402011) is summarized below.
What have we done so far? We collected mature Arabidopsis pollen using the vacuum method and extracted the proteins using three different methods (salt soluble, salt insoluble and SDS soluble). The proteins were separated by 2-dimensional gel electrophoresis (2-DE), the protein spots were excised from the gel and digested with trypsin. The sequences of the resulting peptides were identified by electrospray ionization tandem mass spectrometry (ESI-MS/MS). With the data collected, we generated a proteome reference map of mature Arabidopsis pollen that includes 179 proteins, of which 135 are unique (i.e. have different At numbers).
What did we find? About half of the proteins we identified are involved in metabolism (20%), energy generation (17%) or cell structure (12%); these percentages are similar to those determined for the pollen transcriptome and this similarity is consistent with the idea that in addition to the mRNAs, the mature pollen grain contains proteins necessary for germination and rapid pollen tube growth. We also identified 10 proteins of unknown function, 3 of which are flower or pollen-specific, and we identified 9 proteins whose RNAs were absent from the transcriptome, 7 of which are involved in metabolism, energy generation or cell wall structure.
Of the 9 proteins whose mRNAs are missing from the transcriptome [1, 2, EST representation (NCBI, http://www.ncbi.nlm.nih.gov/)] indicates that none are pollen-specific. For the proteins that are predicted to be secreted, one explanation for their presence might be that they were deposited on the pollen grain by the tapetum, the sporophytic tissue of the anther that surrounds the male gametophytes during their early development. Alternately, for any of these proteins, the mRNA encoding them might be short-lived or be of extremely low abundance and thus appear absent from the transcriptome. Most (7 of 9) of the proteins encoded by mRNAs that are absent from the transcriptome are important in cell structure, energy generation or metabolism. This further supports the idea that the mature pollen grain contains the proteins necessary for germination and rapid pollen tube growth (3).
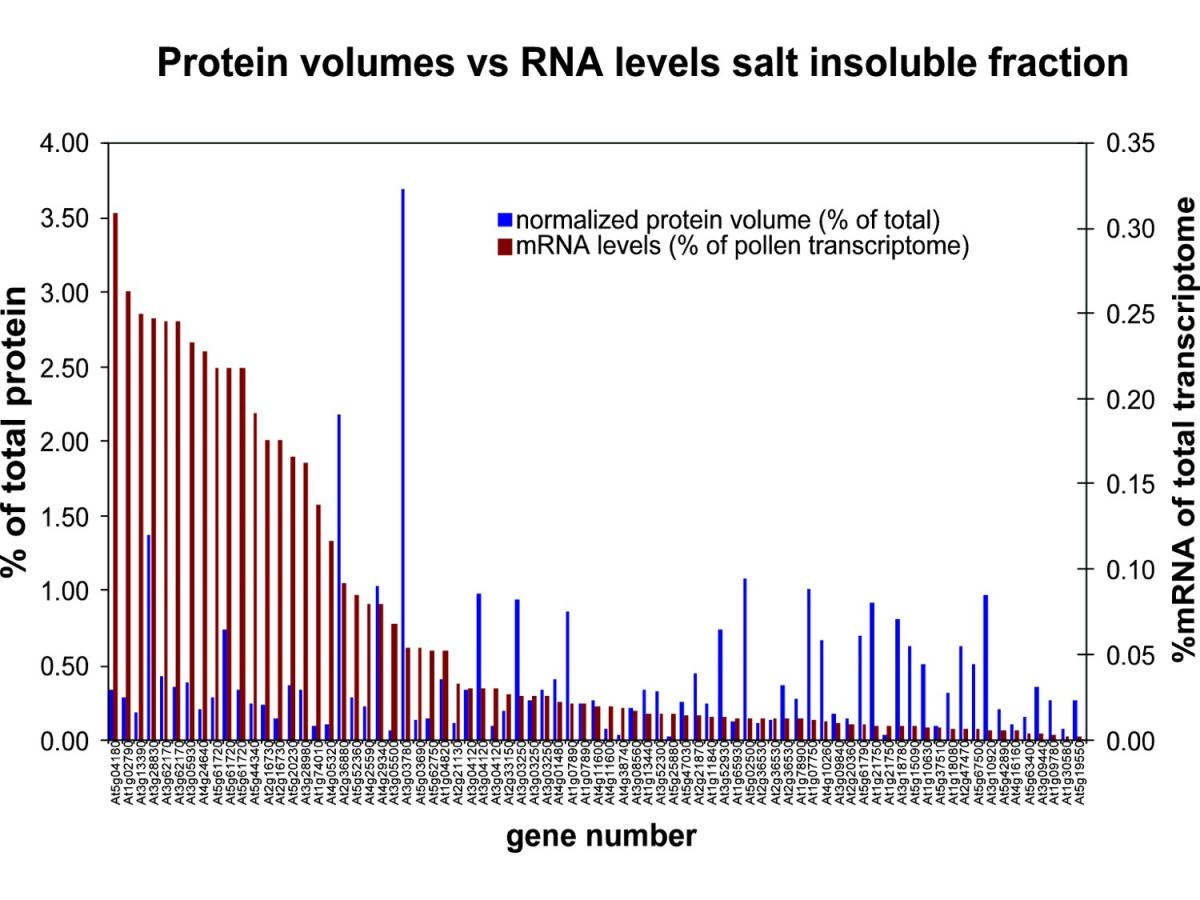
One major limitation to using 2-DE gels for a proteomics study is that only higher abundance proteins are identified. We were interested in knowing whether the more abundant proteins identified also had higher mRNA levels in the transcriptome. To address this question, we quantified the protein spots in all three fractions and compared the normalized protein volumes to their corresponding mRNA signals in the transcriptome 1, 2. We included 77 proteins in this analysis. We excluded those proteins that were identified from spots with multiple identifications and those whose corresponding mRNAs were absent from the mature pollen transcriptome. In order to make comparisons with protein levels, we similarly represented mRNA values as a percentage of the total, using the approximately 6600 genes that are represented in the mature pollen transcriptome 1, 2. In general, we found an inverse relationship: mRNAs that were abundant corresponded to lower abundance proteins while mRNAs that were less abundant corresponded to some of the more abundant proteins. The abundant proteins that had an inverse relationship with their corresponding mRNAs were primarily involved in energy generation, while the low abundance proteins whose mRNAs were abundant were mainly involved in cell structure. It seems reasonable that the mature pollen grain stores proteins it will need for the immediate generation of energy to initiate germination and pollen tube growth and that mRNAs encoding proteins necessary for cell structure are ready for immediate translation upon germination of the pollen grain.
There were, however, exceptions to the inverse relationship: some abundant proteins had corresponding mRNAs that were also high, and some low abundance proteins had corresponding mRNAs that were also low. To determine if there was a trend within this category, we compared the 25 most abundant proteins with the 25 most abundant mRNAs (of those that correspond to proteins we identified) and found that 11 of the 25 mRNAs represented genes important for cell structure, whereas 10 of the 25 proteins are involved in energy production and metabolism. We found only 8 genes whose proteins and mRNAs both fell into the most abundant categories; all are involved in cell structure or metabolism. Among the 25 least abundant proteins and 25 least abundant mRNAs there were 10 genes whose proteins and mRNAs both fell into the least abundant categories. Most of these proteins are necessary for energy generation and signal transduction. Interestingly, the low abundance proteins and mRNAs contained relatively few (three) proteins or mRNAs (one) necessary for cell structure.
The data from this study strongly support the theory that the mature pollen grain puts the majority of its resources into making proteins and mRNAs that it will need immediately upon germination.
Future directions
By establishing this map, we have contributed to the understanding of the biochemistry of pollen and as techniques for in vitro Arabidopsis pollen germination improve (4), comparative proteomic studies between mature pollen and germinated or mutant pollen will become possible. With the ability to isolate relatively large quantities of Arabidopsis pollen, proteomic analyses of sperm (5, 6) should be feasible.
1. AtGenExpress (2004) NASCArrays Experiment: AtGenExpress: Developmental series (flowers and pollen).
2. Craigon, D.J., et al. (2004) NASCArrays: a repository for microarray data generated by NASC's transcriptomics service. Nucleic Acids Res 32 Database issue, D575-577
3. Mascarenhas, J.P. (1975) The biochemistry of angiosperm pollen development. Bot. Rev. 41, 259-314
4. Johnson-Brousseau, S.A., and McCormick, S. (2004) A compendium of methods useful for characterizing Arabidopsis pollen mutants and gametophytically- expressed genes. Plant J 39, 761-775
5. Engel, M.L., et al. (2003) Sperm cells of Zea mays have a complex complement of mRNAs. Plant J 34, 697-707
6. Engel, M.L., et al. (2005) Green Sperm. Identification of male gamete promoters in Arabidopsis thaliana. Plant Physiol 138, 2124-2133.
