University of California, Berkeley | Plant & Microbial Biology
DColeman-Derr

- Principal Investigator
Our laboratory investigates the effects of drought and other abiotic stresses on the microbiomes associated with Sorghum bicolor and other grass species. With the world population expected to reach 9 billion by 2050, it is estimated that the global food supply will need to increase by 70 percent to meet rapidly rising demand. Changes in the global climate may well compound this challenge, as predicted increases in environmental stresses, such as drought and high-salinity, are expected to reduce crop productivity. Recent studies demonstrated that microbial symbionts of crop plants are capable of enhancing the abiotic stress tolerance of their host. However, only a tiny fraction of plant microbiomes have been uncovered and evaluated. Thus, research and new tools are needed to develop a better understanding of the interrelationship between crop plant abiotic stress tolerance and crop plant microbiomes.
Additionally, the lab works on developing statistical methods and computational tools based on genome wide association studies (GWAS) that incorporate epigenomic, transcriptomic, and other data types. In addition to offering increasing the statistical power of GWAS analyses, this approach also has the potential to reveal instances where phenotypic variation is not explained by changes in nucleotide sequence, but instead by epigenetic changes (commonly referred to as “missing heritability”). Additionally, by integrating multiple ‘omics’ datasets, this strategy offers an opportunity to detect correlations among data sets and types and to directly interrogate the complex cascade of signals and forces that leads to phenotype.
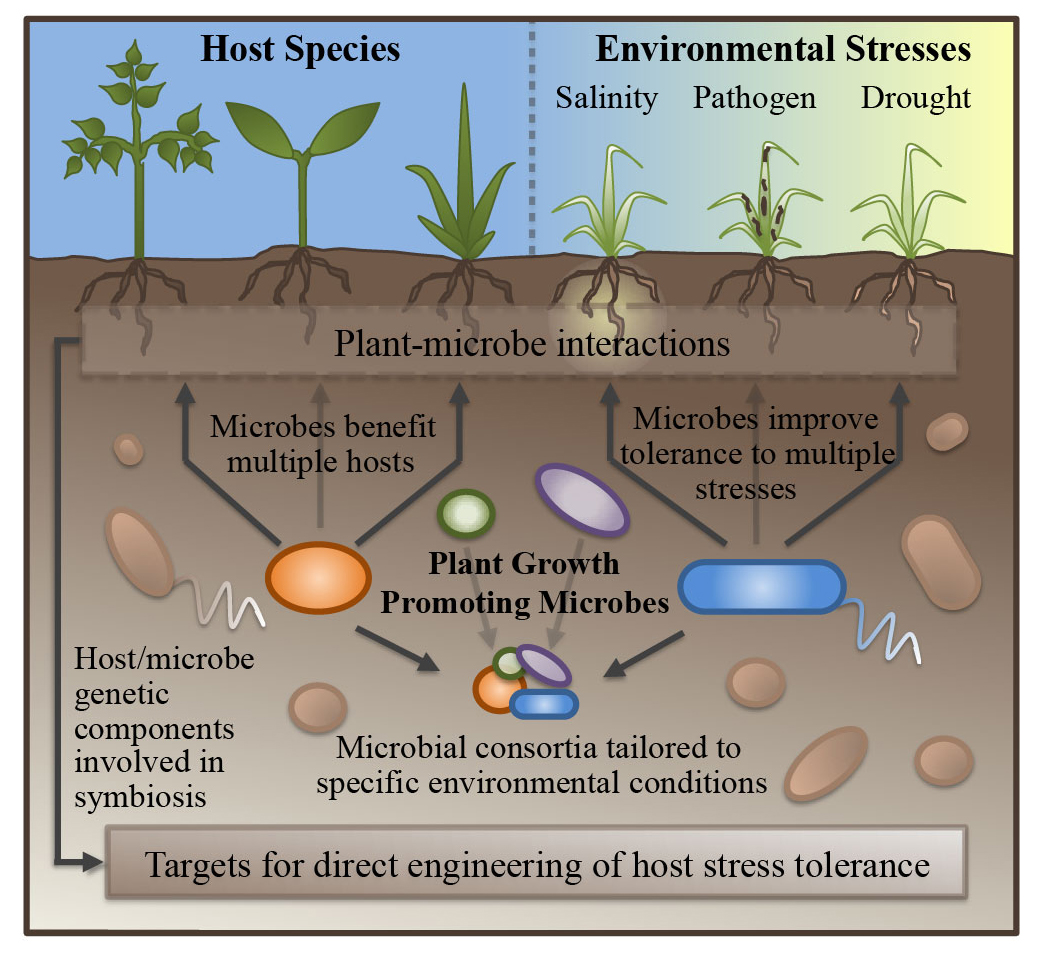
Our lab is also interested in understanding the role of plant growth promoting microbes (PGPM) in improving abiotic stress response in their plant hosts. Using a variety of culture and genomics-based approaches, we are working to uncover the host machinery manipulated by several PGPM species, which in turn may serve as potential targets for direct crop improvement through conventional breeding practices.
Educational Background:
Ph.D. Plant Biology University of California at Berkeley, 2012
B.A. Natural Sciences New College of Florida, 1998
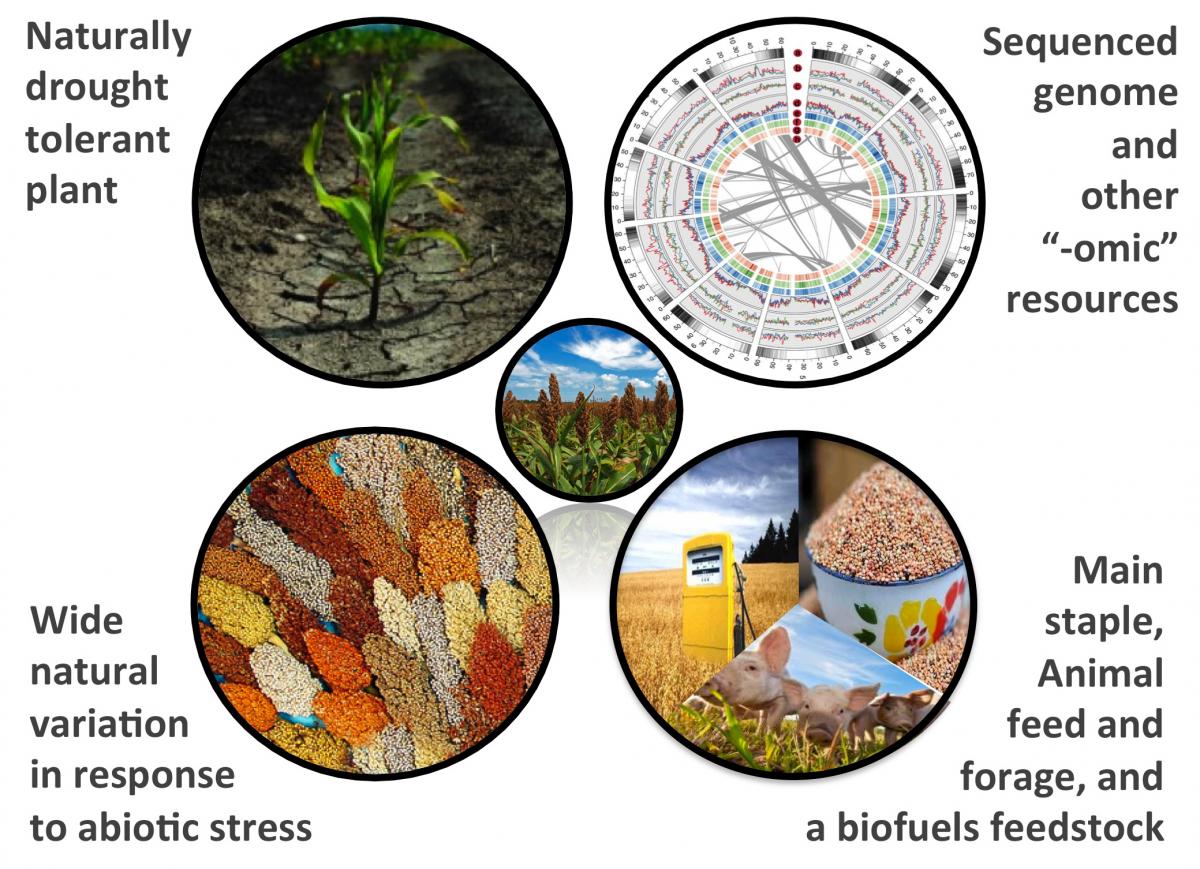
Natural variaton in drought response in Sorghum bicolor and other crop species
Drought stress is arguably the most critical factor for crop production in many parts of the world and will increasingly become a greater factor with climate change; many microbial species have been shown to be capable of improving drought response in a variety of crop species. Sorghum bicolor, a crop with both feedstock and biofuels potential, is frequently grown in dry climates with frequent periods of drought. Recently, a genotype-by-sequencing (GBS) approach was used to generate a large SNP dataset for a large Sorghum association panel; these data and germplasm are publically available. Our research involves generating experimentally linked epigenetic and transcriptomic datasets using these accessions to investigate the correlation between genomic, epigenomic and transcriptomic response under drought stress. A variety of drought response phenotypes, including root morphology, biomass, and chlorophyll content are being used to detect associations between genetic variation in this broad panel of Sorghum lines and their ability to thrive in environments with limited water availability.
Sorghum as a model for drought tolerance research
Plant growth promoting microbes and abiotic stress response in crop species
A large variety of plant growth promoting microbes (PGPM) have been shown to be capable of reducing abiotic stress response in their plant hosts, though the mechanisms through which they act are still being explored and established. In addition to a number of characterized strategies of growth promotion, including improved nutrient exchange, production of phytohormones, and competition with pathogens, recent research continues to unveil novel pathways through which PGPM benefit their hosts. Using genomics-based approaches, we are working to uncover the host machinery manipulated by several PGPM species in Sorghum and other species.
Mechanisms of plant growth promotion by microbial symbionts